|
|
MEPSAnd: Minimum Energy Path Surface Analysis over n-dimensional surfaces.
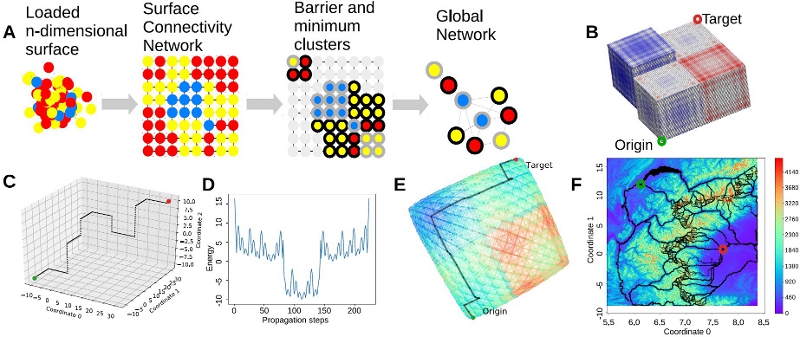
n-dimensional energy surfaces are becoming computationally accessible, yet interpreting their information is not straightforward. We present MEPSAnd, an open source GUI-based program that natively calculates minimum energy paths across energy surfaces of any number of dimensions. Among other features, MEPSAnd can compute the path through lowest barriers and automatically provide a set of alternative paths. MEPSAnd offers distinct plotting solutions as well as direct python scripting.
Availability and implementation: MEPSAnd is freely available (under GPLv3 license) at this page: [http://bioweb.cbm.uam.es/software/MEPSAnd/]
CITATION: Marcos-Alcalde, I., L-V, E. & Gómez-Puertas, P. (2020). MEPSAnd: Minimum Energy Path Surface Analysis over n-dimensional surfaces. Bioinformatics 36, 956–958. [Free-access article link] [https://doi.org/10.1093/bioinformatics/btz649]
|
|
|
MEPSAnd DOWNLOAD (freely available, under GPLv3 license):
MEPSAnd v1.6 (Mar. 2022):
MEPSAnd v1.6 source code: [MEPSAnd_v1.6.zip]
MEPSAnd v1.6 user manual (42.5 MB): [MEPSAnd_manual_v1.6.zip]
DATA SAMPLES (MEPSAnd user manual): [examples.zip]
New in version 1.6:
- New "Assume grid" connectivity prediction mode that significantly accelerates the neighboring calculations for homogeneously distributed datasets (the most common use case).
- Fixed subtle bug with global node ids annotation that could produce an error when selecting non-minimum origin or target points and could block network plots from working with networks containing disconnected regions.
- Added a check for the existence of disconnected regions that reports a warning message.
- Added a check for coordinate redundancy to prevent the use of artefactual surfaces.
- Thanks to Dr. Marcelo D. Polęto's feedback, an automatic data type reduction function to reduce memory usage was implemented.
- Thanks to Dr. Marcelo D. Polęto's feedback, the connectivity calculations were rewritten to reduce memory usage.
- Changed the way fragmentwise path is saved to avoid extensive memory use when extremely complex paths are calculated.
- Major overhaul of the "Energy profiles" and "Coordinate projections" point labeling logic. Now only minimum or barrier points are labeled, indicating the cluster number to which they belong.
- Made it possible to plot "Coordinate projections" without choosing any point.
- MEPSAnd no longer requires igraph, ForceAtlas2 or cairo to run. If any of these packages is not detected, "Network projections" will be disabled but everything else will work as usual.
- Replaced "dype=np.int" instances with "dtype=np.int_".
- Added terminal report messages indicating the number of neighbors found as a quick sanity check for connectivity calculations.
- Added terminal report messages indicating minima propagation progress.
- Removed instances of fig.gca(projection='3d') as it is deprecated matplotlib.
- Instances of "map_" in the source code have been replaced with "surface_" to unify terminology.
- Fixed well sampling calculations not identifying origin or target clusters correctly.
- Minor code cleaning.
- A "Good practices" section has been added to the manual, with a particular focus on metadynamics, based on Dr. Marcelo D. Polęto's reports.
FORMER VERSIONS (no longer supported):
MEPSAnd v1.41 (Jul. 2020):
MEPSAnd v1.41 source code: [MEPSAnd_v1.41.zip]
MEPSAnd v1.41 user manual (higher resolution -43 MB- ): [MEPSAnd_manual_v1.41_l.zip]
MEPSAnd v1.41 user manual (lower resolution -3 MB- ): [MEPSAnd_manual_v1.41_s.zip]
DATA SAMPLES (MEPSAnd user manual): [examples.zip]
New in version 1.41:
- Added the correct citation reference to MEPSAnd.py, replacing the previous "XXXX..." placeholder.
- Updated the citation reference in the manual, as the paper is no longer "in press".
MEPSAnd v1.4 (Nov. 2019):
MEPSAnd v1.4 source code: [MEPSAnd_v1.4.zip]
MEPSAnd v1.4 user manual: [MEPSAnd_manual_v1.4.zip]
New in version 1.4:
- Number formatting in "save_simplified_path_to_txt" method has been modified to improve the readability of the resulting files.
MEPSAnd v1.35 (Nov. 2019):
MEPSAnd v1.35 source code: [MEPSAnd_v1.35.zip]
New in version 1.35:
- BUGFIX: a bug introduced during network projection rewriting in 1.3 broke the ability to properly assign default plotting parameters in "barriers as vertices" graphs.
MEPSAnd v1.3 (Nov. 2019):
MEPSAnd v1.3 source code: [MEPSAnd_v1.3.zip]
New in version 1.3:
- Major fix: network projection edge selection was not working as intended and has been completely rewritten.
- Minor bugfixes to the GUI.
- Minor code cleaning.
MEPSAnd v1.2 (Oct. 2019):
MEPSAnd v1.2 source code: [MEPSAnd_v1.2.zip]
New in version 1.2:
- BUGFIX: As reported by Oriol Esquivias, the method to save minimum and barrier clusters was broken when clusters of more than one point were present. Fixed using an index array ("index_array") with the adequate shape in the "np.column_stack" calls of the "save_minbar_clusters_to_txt" method.
- Changed how the MEPSAnd version is printed on the terminal when it is launched to improve version readability.
MEPSAnd v1.1 (Sep. 2019):
MEPSAnd v1.1 source code: [MEPSAnd_v1.1.zip]
New in version 1.1:
- BUGFIX: As reported by Oriol Esquivias, instances of pandas compat.StringIO could lead to incompatibility issues with pandas 0.25.1.
Thus, all instances of pd.compat.StringIO have been replaced with io.StringIO.
MEPSAnd v1.0 (Aug. 2019):
|
|
|
|
|
BIOWEB: MOLECULAR MODELLING GROUP
Centro de Biología Molecular "Severo Ochoa" (CSIC-UAM).
http://bioweb.cbm.uam.es |
C/ Nicolás Cabrera, 1. Campus UAM, Cantoblanco.
28049 Madrid. Spain
Tel: (+34) 91 196 4662 / 4663
Fax: (+34) 91 196 4420
|
|
|
|